Application progress of artificial intelligence in the analysis of tumor-tissue histopathological biomarkers
-
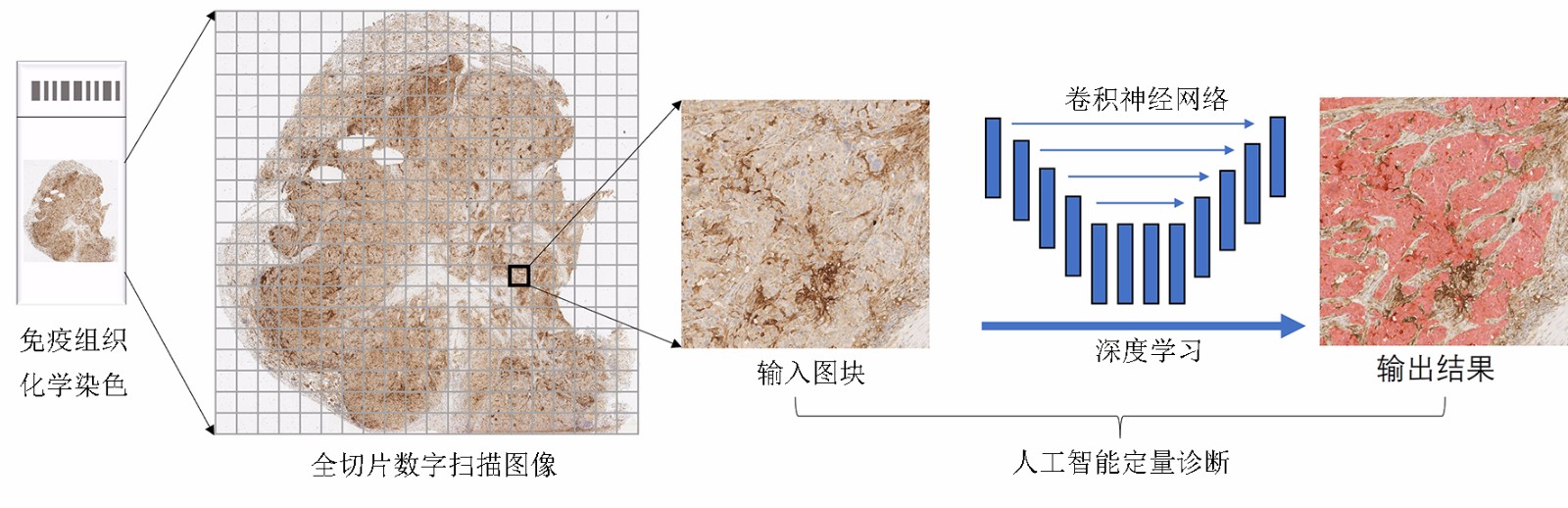
摘要: 肿瘤个体化治疗的进展对肿瘤组织病理标记物的精确诊断提出了更高的要求。数字病理(digital pathology,DP)的发展为人工智能(artificial intelligence,AI)辅助诊断在肿瘤组织病理图像分析中的应用提供了基础。基于卷积神经网络的深度学习(deep learning,DL)算法能够将DP图像与计算机分析技术相结合,有望成为定量评价肿瘤组织生物标志物的重要工具。本文概述了AI在组织病理学中的发展,并以目前研究相对广泛且与临床诊疗密切相关的分子病理指标Her-2、Ki-67及PD-L1的图像分析为具体案例,重点阐述了当前AI在肿瘤病理标志物分析中的研究进展。AI辅助的肿瘤病理诊断具有客观性强及可重复性高等优点,能够实现肿瘤组织标志物诊断的定量分析,从而克服病理医生人工判读的挑战,提高病理诊断的精确性。通过计算机工具构建肿瘤组织标志物的AI判读模式,是构建未来肿瘤智能诊疗体系的重要环节。Abstract: The progress of tumor individualized treatment puts forward higher requirements for the accurate diagnosis of tumor tissue-based specific biomarkers. The development of digital pathology provides a basis for the application of artificial intelligence (AI)-assisted diagnostic tools in tumor pathological image analysis. The deep learning algorithm based on a convolutional neural network combines digital pathological images with computer analysis technology, which is an important tool for quantitative evaluation of tumor-tissue biomarkers. This review summarizes the development of AI in histopathology and focuses on the progress of image analysis of molecular biomarkers such as Her-2, Ki-67, and PD-L1. These molecular biomarkers are specificly supported by extensive research and closely related to clinical diagnosis and treatment. Studies have shown that AI-assisted tumor diagnosis has the advantages of strong objectivity and high repeatability. It can obtain the quantitative results of the tumor-tissue biomarkers to overcome the challenges of manual interpretation and improve the accuracy of diagnosis of pathologists. The development of AI-based analysis tools of tumor-tissue biomarkers is an important method to build intelligent and accurate tumor diagnosis and treatment systems of the future.
-
Key words:
- artificial intelligence (AI) /
- digital pathology /
- tumor biomarkers
-
表 1 PD-L1评分AI判读模型的相关研究
文献 肿瘤类型 抗体克隆 表达评分 学习模式 AI模型与人工评分的比较 Koelzer等[23] 黑色素瘤 SP263 肿瘤细胞评分 随机森林树/监督学习 r=0.97,P<0.000 1 Kapil等[24] 非小细胞肺癌 SP263 肿瘤细胞阳性比例分数(TPS) 生成对抗网络/半监督学习 OPA=0.88;NPA=0.88,PPA=0.85;
Lin's CCC=0.94;Pearson CCC=0.95;Taylor等[25] 非小细胞肺癌 22c3 肿瘤细胞评分;
免疫细胞评分带反馈回路的监督学习 %TC:Lin's CCC=0.68~0.81;
%IC:Lin's CCC=0 0.53~0.88;Wu等[26] 非小细胞肺癌 22c3;SP263 肿瘤细胞阳性比例分数(TPS) U-ResNet/监督学习 22c3:r=0.942 9~0.945 8;
SP263:r=0.978 7AI:人工智能;OPA:总符合率;NPA:阴性符合率;PPA:阳性符合率;Lin's CCC:Lin的一致性相关系数;Pearson CCC:皮尔逊一致性相关系数;%TC:肿瘤细胞阳性比例;%IC:免疫细胞阳性比例 -
[1] Pignatti F, Ehmann F, Hemmings R, et al. Cancer drug development and the evolving regulatory framework for companion diagnostics in the EuropeanUnion [J]. Clin Cancer Res, 2014, 20(6):1458-1468. [2] Bera K, Schalper KA, Rimm DL, et al. Artificial intelligence in digital pathology-new tools for diagnosis and precision oncology[J]. Nat Rev Clin Oncol, 2019, 16(11):703-715. doi: 10.1038/s41571-019-0252-y [3] LeCun Y, Bengio Y, Hinton G. Deep learning [J]. Nature, 2015, 521(7553):436-444. [4] Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence[J]. Lancet Oncol, 2019, 20(5):253-261. doi: 10.1016/S1470-2045(19)30154-8 [5] Tuominen VJ, Tolonen TT, Isola J. ImmunoMembrane: a publicly available web application for digital image analysis of HER2 immunohistochemistry[J]. Histopathology, 2012, 60(5):758-767. doi: 10.1111/j.1365-2559.2011.04142.x [6] Brugmann A, Eld M, Lelkaitis G, et al. Digital image analysis of membrane connectivity is a robust measure of HER2 immunostains[J]. Breast Cancer Res Treat, 2012, 132(1):41-49. doi: 10.1007/s10549-011-1514-2 [7] Saha M, Chakraborty C. Her2Net: A Deep framework for semantic segmentation and classification of cell membranes and nuclei in breast cancer evaluation [J]. IEEE Trans Image Process, 2018, 27(5):2189-2200. [8] Qaiser T, Mukherjee A, Reddy PBC, et al. HER2 challenge contest: a detailed assessment of automated HER2 scoring algorithms in whole slide images of breast cancer tissues[J]. Histopathology, 2018, 72(2):227-238. doi: 10.1111/his.13333 [9] Tewary S, Arun I, Ahmed R, et al. AutoIHC-Analyzer: computer-assisted microscopy for automated membrane extraction/scoring in HER2 molecular markers[J]. J Microsc, 2021, 281(1):87-96. doi: 10.1111/jmi.12955 [10] Hofener H, Homeyer A, Forster M, et al. Automated density-based counting of FISH amplification signals for HER2 status assessment[J]. Comput Methods Programs Biomed, 2019, 173:77-85. doi: 10.1016/j.cmpb.2019.03.006 [11] Yue M, Zhang J, Wang X, et al. Can AI-assisted microscope facilitate breast HER2 interpretation? A multi-institutional ring study[J]. Virchows Arch, 2021, 479(3):443-449. doi: 10.1007/s00428-021-03154-x [12] Vandenberghe ME, Scott ML, Scorer PW, et al. Relevance of deep learning to facilitate the diagnosis of HER2 status in breast cancer[J]. Sci Rep, 2017, 7:45938. doi: 10.1038/srep45938 [13] Kloppel. Pancreatic neuroendocrine tumors: update on the new World Health Organization classification[J]. Ajsp-Rev Rep, 2018, 23(4):193-194. [14] Goodell PP, Krasinskas AM, Davison JM, et al. Comparison of methods for proliferative index analysis for grading pancreatic well-differentiated neuroendocrine tumors[J]. Am J Clin Pathol, 2012, 137(4):576-582. doi: 10.1309/AJCP92UCXPJMMSDU [15] Voros A, Csorgo E, Nyari T, et al. An intra- and interobserver reproducibility analysis of the Ki-67 proliferation marker assessment on core biopsies of breast cancer patients and its potential clinical implications[J]. Pathobiology, 2013, 80(3):111-118. doi: 10.1159/000343795 [16] 郭蕾,石峰,郑闪等.乳腺癌Ki-67计算机全自动分析可行性探讨[J].中华肿瘤防治杂志,2015,22(9):678-681. [17] Dirican E, Kilic E. A machine learning approach for the association of ki-67 scoring with prognostic factors [J]. J Oncol, 2018, 7:1912438. [18] Wang HY, Li ZW, Sun W, et al. Automated quantification of Ki-67 index associates with pathologic grade of pulmonary neuroendocrine tumors[J]. Chinese Med J-Peking, 2019, 132(5):551-561. doi: 10.1097/CM9.0000000000000109 [19] Niazi MKK, Tavolara TE, Arole V, et al. Identifying tumor in pancreatic neuroendocrine neoplasms from Ki67 images using transfer learning[J]. Plos One, 2018, 13(4):e0195621. doi: 10.1371/journal.pone.0195621 [20] Valkonen M, Isola J, Ylinen O, et al. Cytokeratin-supervised deep learning for automatic recognition of epithelial cells in breast cancers stained for ER, PR, and Ki-67[J]. IEEE Trans Med Imaging, 2020, 39(2):534-542. doi: 10.1109/TMI.2019.2933656 [21] Cooper WA, Russell PA, Cherian M, et al. Intra- and interobserver reproducibility assessment of PD-L1 biomarker in non-small cell lung cancer[J]. Clin Cancer Res, 2017, 23(16):4569-4577. doi: 10.1158/1078-0432.CCR-17-0151 [22] Brunnstrom H, Johansson A, Westbom-Fremer S, et al. PD-L1 immunohistochemistry in clinical diagnostics of lung cancer: inter-pathologist variability is higher than assay variability[J]. Mod Pathol, 2017, 30(10):1411-1421. doi: 10.1038/modpathol.2017.59 [23] Koelzer VH, Gisler A, Hanhart JC, et al. Digital image analysis improves precision of PD-L1 scoring in cutaneous melanoma[J]. Histopathology, 2018, 73(3):397-406. doi: 10.1111/his.13528 [24] Kapil A, Meier A, Zuraw A, et al. Deep semi supervised generative learning for automated tumor proportion scoring on NSCLC tissue needle biopsies[J]. Sci Rep-Uk, 2018, 8(1):17343. [25] Taylor CR, Jadhav AP, Gholap A, et al. A multi-institutional study to evaluate automated whole slide scoring of immunohistochemistry for assessment of programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer[J]. Appl Immunohistochem Mol Morphol, 2019, 27(4):263-269. doi: 10.1097/PAI.0000000000000737 [26] Wu J, Liu C, Liu X, et al. Artificial intelligence-assisted system for precision diagnosis of PD-L1 expression in non-small cell lung cancer[J]. Mod Pathol, 2021,35(3):403-411. [27] Wu J, Lin D. A review of artificial intelligence in precise assessment of programmed cell death-ligand 1 and tumor-infiltrating lymphocytes in non-small cell lung cancer[J]. Adv Anat Pathol, 2021, 28(6):439-445. [28] 金昱,潘凯枫,张艺宝,等.人工智能在肿瘤三级预防中的应用机遇与挑战[J].中国肿瘤临床,2021,48(21):1081-1087. [29] Zhang J, Song J, Sheng J, et al. Multiplex imaging reveals the architecture of the tumor immune microenvironment[J]. Cancer Biol Med, 2021, 18(4):949-954. doi: 10.20892/j.issn.2095-3941.2021.0494 [30] Koelzer VH, Sirinukunwattana K, Rittscher J, et al. Precision immunoprofiling by image analysis and artificial intelligence[J]. Virchows Arch, 2019, 474(4):511-522. doi: 10.1007/s00428-018-2485-z [31] Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images[J]. Nat Med, 2019, 25(8):1301-1309. doi: 10.1038/s41591-019-0508-1 [32] Budd S, Robinson EC, Kainz B. A survey on active learning and human-in-the-loop deep learning for medical image analysis[J]. Med Image Anal, 2021, 71:102062. doi: 10.1016/j.media.2021.102062 [33] Cheng JY, Abel JT, Balis UGJ, et al. Challenges in the development, deployment, and regulation of artificial intelligence in anatomic pathology[J]. Am J Pathol, 2021, 191(10):1684-1692. doi: 10.1016/j.ajpath.2020.10.018 [34] Jiang Y, Yang M, Wang S, et al. Emerging role of deep learning-based artificial intelligence in tumor pathology[J]. Cancer Commun (Lond), 2020, 40(4):154-166. doi: 10.1002/cac2.12012 -


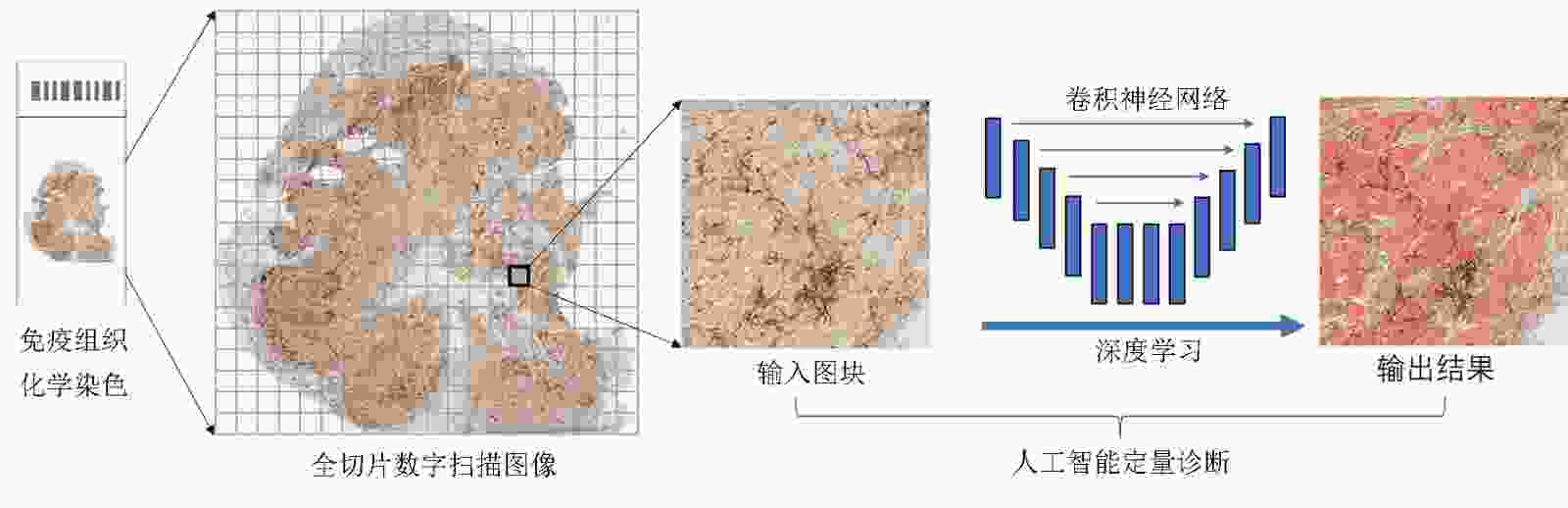
 下载:
下载: