-
摘要:
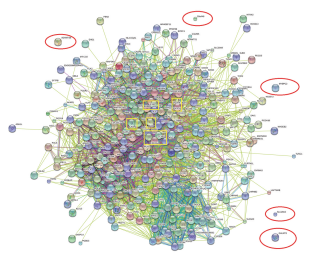
目的 筛选肺癌患病风险相关的关键基因, 为进一步的相关性研究提供参考。 方法 基于文献挖掘的方法统计出肺癌患病风险相关基因, 运用生物信息学方法对上述基因进行分析。 结果 近10年内的文献共涉及312个基因, 其中不同分级的GO分类762种, KEGG通路81种, 而ln大于0且P值≤0.001的GO分类有70种, KEGG通路8种, 两者主要与细胞生理、代谢、增殖、DNA损伤修复及细胞凋亡、细胞周期、MAPK信号通路有关。在线String软件构建的蛋白网络图提示这些基因的表达产物大部分之间存在着密切的相互作用关系, 同时发现上述基因中307个可翻译成蛋白并参与网络构建。进一步运用Cytoscape软件将"String"软件所构建的蛋白网络图可视化及量化同时筛选出24个关键基因。 结论 肺癌患病风险相关基因种类繁多, 针对关键基因的大样本跨种族高质量肺癌患病风险临床研究是未来的研究方向。 Abstract:Objective This study aimed to determine the key genes that are associated with lung cancer risk for further study. Methods Techniques in bioinformatics were applied in lung cancer risk genes that were obtained from clinical trials. Results We obtained 312 lung cancer risk genes that are mainly related to cellular physiological processes, metabolism, cell proliferation, DNA repair and cell cycle, apoptosis, and MAPK signaling pathway. A total of 762 functional classifications by the Gene Ontology (GO) and 81 pathways by the Kyoto _Encyclopedia of Genes and Genomes (KEGG) were involved. Among these risk genes, only 70 GO classifications and 8 KEGG pathways were significant [ln(Bayes factor)≥0 and P-value≤0.001]. Only 307 genes can be translated into proteins; thus, the complex pro tein-protein network that was built using the Online String software also revealed 307 nodes. This network was visualized and quantified using the Cytoscape software. We determined 24 key genes from the protein-protein network. Conclusion Selecting the key genes from a large number of lung cancer risk genes and performing a large sample interracial clinical study are important in the analysis of lung cancer key risk genes through bioinformatics. -
Key words:
- Lung cancer /
- Risk /
- Key genes /
- Bioinformatics
-
表 1 肺癌患病风险相关基因主要GO功能分类
Table 1. Major gene ontology classification of proteins that were expressed by the lung cancer risk genes

表 2 肺癌患病风险相关基因涉及的KEGG通路
Table 2. Pathways of proteins that were expressed by the lung cancer risk genes in the Kyoto Encyclopedia of Genes and Genomes

表 3 肺癌患病风险相关的24个关键候选基因表
Table 3. Twenty-four key candidate genes of lung cancer

-
[1] Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008[J]. Int J Cancer, 2010, 127(12): 2893-2917. doi: 10.1002/ijc.25516 [2] Bruske-Hohlfeld I. Environmental and occupational risk factors for lung cancer[J]. Methods Mol Biol, 2009, 472: 3-23. [3] Brenner DR, McLaughlin JR, Hung RJ. Previous lung diseases and lung cancer risk: a systematic review and meta-analysis[J]. PLoS One, 2011, 6(3): e17479. doi: 10.1371/journal.pone.0017479 [4] Yin J, Vogel U, Ma Y, et al. HapMap-based study of the DNA repair gene ERCC2 and lung cancer susceptibility in a Chinese population[J]. Carcinogenesis, 2009, 30(7): 1181 -1185. doi: 10.1093/carcin/bgp107 [5] Truong T, Sauter W, McKay JD, et al. International Lung Cancer Consortium: co-ordinated association study of 10 potential lung cancer susceptibility variants[J]. Carcinogenesis, 2010, 31(4): 625-633. doi: 10.1093/carcin/bgq001 [6] Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomicsignatures[J]. Bioinformatics, 2006, 22(23): 2926-2933. doi: 10.1093/bioinformatics/btl483 [7] Szklarczy D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored[J]. Nucleic Acids Res, 2011, 39(Database issue): D561-568. [8] Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks[J]. Methods Mol Biol, 2011, 696: 291-303. [9] Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameterswith CentiScaPe[J]. Bioinformatics, 2009, 25(21): 2857-2859. doi: 10.1093/bioinformatics/btp517 [10] Risch A, Plass C. Lung cancer epigenetics and genetics[J]. Int J Cancer, 2008, 123(1): 1-7. doi: 10.1002/ijc.23605 [11] Kiyohara C, Yoshimasu K, Takayama K, et al. Lung cancer susceptibility: are we on our way to identifying a high-risk group[J]. Future Oncol, 2007, 3(6): 617-627. doi: 10.2217/14796694.3.6.617 [12] Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium[J]. Nat Genet, 2000, 25(1): 25-29. doi: 10.1038/75556 [13] Marden JI. Hypothesis testing: From values to Bayes. factors[J]. J Am Statist Assoc, 2000, 95(452): 1316-1320. [14] Yang M, Choi Y, Hwangbo B, et al. Combined effects of genetic polymorphisms in six selected genes on lung cancer susceptibility[J]. Lung Cancer, 2007, 57(2): 135-142. doi: 10.1016/j.lungcan.2007.03.005 [15] Honma HN, De Capitani EM, Perroud MW Jr, et al. Influence of p53 codon 72 exon 4, GSTM1, GSTT1 and GSTP1* B polymorphisms in lung cancer risk in a Brazilian population[J]. Lung Cancer, 2008, 61(2): 152-162. doi: 10.1016/j.lungcan.2007.12.014 -



 下载:
下载: